Conservation Genetics; Wolf tortoises with Floreana (extinct) genes.
The volcano Wolf tortoise population (Chelonoidis becki) is a bag of surprises.
Previous work by Michael Russello (British Columbia University) showed that some unmarked individuals from Volcano Wolf in Northern Isabela had shared ancestry with C. elephantopus, a now, extinct species from Floreana Island.
An expedition was put together by Dr. Gisella Caccone, and +1669 tortoises from Volcano Wolf were, sexed, tagged and blood samples taken.
Here is when Ryan Garrick (now at Ole' Miss.) and me, enter the picture. We completed a reference data set adding more genotypes from Floreana Museum samples (this was done in a dedicated facility for the extraction of historical DNA from bone remains), and we genotyped 12 microsatellite loci in 1669 individuals from Volcano Wolf.
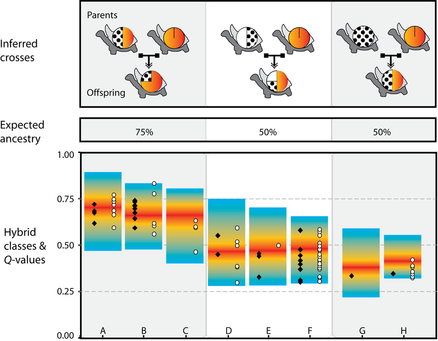
We compared these 1669 Wolf individuals with a reference data set containing all extant and extinct Galapagos tortoises to look out for Wolf tortoises with Floreana ancestry. We were also interested in estimating how much of each individual genome was traceable to each of the parent lineages. The figure on top shows individuals with mixed ancestry allotted in eight simulated hybridization classes. Our final results identified 84 Wolf individuals that seem to be immediate descendants of a purebred C. elephantopus (Floreana). This result was further substantiated by showing that out of 84 tortoises, 26 of them also had a C. elephantopus-like mitochondrial haplotype.
There are important implications to this finding. For example, Floreana (like the rest of Galapagos islands) is an ecosystem that has evolved with the presence of Tortoises as the main herbivores. In effect, there is strong evidence showing that tortoises act as ecosystem engineers, keeping the balance of each island plant communities. With the tortoises gone this balance is broken, facilitating perhaps the success of invasive plants and radically changing the original ecosystem. Islands are fairly sensitive to invasive species (as shown by the Hawaii Islands), they are sensitive ecosystems as a whole.
One way to preserve Galapagos environments is to keep the past community components together, and hence reintroducing tortoises to islands that harbored them in the past might be a key step 'buffering' Galapagos islands ecosystems against invasives.
From an evolutionary genetics perspective, reintroducing tortoises, is akin to reintroduce individual genes. It makes sense to reintroduce genes that have accumulated additive genetic variance resulting from evolutionary processes taking place in Floreana. Hence, the importance of finding tortoises with some degree of shared ancestry to the extinct lineages.
Previous work by Michael Russello (British Columbia University) showed that some unmarked individuals from Volcano Wolf in Northern Isabela had shared ancestry with C. elephantopus, a now, extinct species from Floreana Island.
An expedition was put together by Dr. Gisella Caccone, and +1669 tortoises from Volcano Wolf were, sexed, tagged and blood samples taken.
Here is when Ryan Garrick (now at Ole' Miss.) and me, enter the picture. We completed a reference data set adding more genotypes from Floreana Museum samples (this was done in a dedicated facility for the extraction of historical DNA from bone remains), and we genotyped 12 microsatellite loci in 1669 individuals from Volcano Wolf.
We compared these 1669 Wolf individuals with a reference data set containing all extant and extinct Galapagos tortoises to look out for Wolf tortoises with Floreana ancestry. We were also interested in estimating how much of each individual genome was traceable to each of the parent lineages. The figure on top shows individuals with mixed ancestry allotted in eight simulated hybridization classes. Our final results identified 84 Wolf individuals that seem to be immediate descendants of a purebred C. elephantopus (Floreana). This result was further substantiated by showing that out of 84 tortoises, 26 of them also had a C. elephantopus-like mitochondrial haplotype.
There are important implications to this finding. For example, Floreana (like the rest of Galapagos islands) is an ecosystem that has evolved with the presence of Tortoises as the main herbivores. In effect, there is strong evidence showing that tortoises act as ecosystem engineers, keeping the balance of each island plant communities. With the tortoises gone this balance is broken, facilitating perhaps the success of invasive plants and radically changing the original ecosystem. Islands are fairly sensitive to invasive species (as shown by the Hawaii Islands), they are sensitive ecosystems as a whole.
One way to preserve Galapagos environments is to keep the past community components together, and hence reintroducing tortoises to islands that harbored them in the past might be a key step 'buffering' Galapagos islands ecosystems against invasives.
From an evolutionary genetics perspective, reintroducing tortoises, is akin to reintroduce individual genes. It makes sense to reintroduce genes that have accumulated additive genetic variance resulting from evolutionary processes taking place in Floreana. Hence, the importance of finding tortoises with some degree of shared ancestry to the extinct lineages.
Conservation genetics; Telmatobius culeus the giant frog of lake Titicaca

Telmatobius is the quintessential high andean frog. The genus is completely aquatic, and if you find a Telmatobius below 2000m of elevation. Well, that will be news. I have worked with one of the most charismatic species in the genus; the giant frog of Lake Titicaca (3800m).
This is asingular species, and here I'm using a picture that is a still of a fantastic video (follow the link): (http://www.arkive.org/titicaca-water-frog/telmatobius-culeus/video-10.html).
Apart from a unique external morphology this frog is an endangered species and the IUCN has listed it as a critically endangered species. (http://www.iucnredlist.org/apps/redlist/details/57334/0). This was one of the many reasons behind the decision to start research on them.
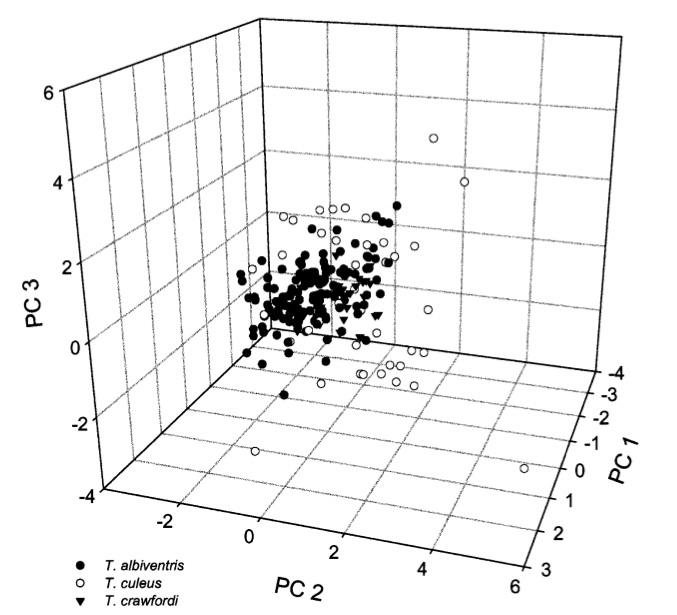
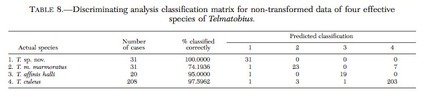
To begin with, the taxonomy of Telmatobius culeus and the many related species was rather obscure and had not been formally revised since the Agassiz and Garman's monography published in 1875 (Not sure if this was Louis Agassiz from the Harvard Museum of Comparative Zoology). One of the obvious problems, was the identity of T. culeus compared to other forms described within or nearby the Lake Titicaca. To solve the issue I used 13 allozyme polymorphisms and classic morphometrics to investigate 'species boundaries' within this species complex. The study supported synonymyzing three species names, luckily keeping the name Telmatobius culeus as a senior synonym. The same data showed discriminatory power with closely related species as evidenced by discriminant functions (Figures below). In this case morphology matched inferences made using allozyme allele frequencies.
More recently, I have used mtDNA sequences with somehow similar results. Mitochondrial DNA is a good 'first pass' indicator of population structure and genetic divergence. However, single locus mitochondrial demographic parameters are limited and not vey accurate. A more correct assessment of key demographic aspects from endangered populations can be drawn from microsatellite loci polymorphisms. In this respect, this type of data needs to be generated in the future.
Similarly, because little is known about the effect of past climate changes in the high andean plateau and its connection to evolutionary processes of Andean endemics in general, it would be nice to add nuclear sequence loci data for a correct understanding of evolutionary and ecological timescales.
This is asingular species, and here I'm using a picture that is a still of a fantastic video (follow the link): (http://www.arkive.org/titicaca-water-frog/telmatobius-culeus/video-10.html).
Apart from a unique external morphology this frog is an endangered species and the IUCN has listed it as a critically endangered species. (http://www.iucnredlist.org/apps/redlist/details/57334/0). This was one of the many reasons behind the decision to start research on them.
To begin with, the taxonomy of Telmatobius culeus and the many related species was rather obscure and had not been formally revised since the Agassiz and Garman's monography published in 1875 (Not sure if this was Louis Agassiz from the Harvard Museum of Comparative Zoology). One of the obvious problems, was the identity of T. culeus compared to other forms described within or nearby the Lake Titicaca. To solve the issue I used 13 allozyme polymorphisms and classic morphometrics to investigate 'species boundaries' within this species complex. The study supported synonymyzing three species names, luckily keeping the name Telmatobius culeus as a senior synonym. The same data showed discriminatory power with closely related species as evidenced by discriminant functions (Figures below). In this case morphology matched inferences made using allozyme allele frequencies.
More recently, I have used mtDNA sequences with somehow similar results. Mitochondrial DNA is a good 'first pass' indicator of population structure and genetic divergence. However, single locus mitochondrial demographic parameters are limited and not vey accurate. A more correct assessment of key demographic aspects from endangered populations can be drawn from microsatellite loci polymorphisms. In this respect, this type of data needs to be generated in the future.
Similarly, because little is known about the effect of past climate changes in the high andean plateau and its connection to evolutionary processes of Andean endemics in general, it would be nice to add nuclear sequence loci data for a correct understanding of evolutionary and ecological timescales.